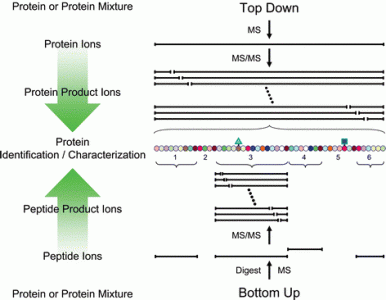
Top Down Mass Spectrometry
Top Down Mass Spectrometry is a technique to characterize the entire sequence of a protein with its related modifications. This service line is tailored to the comprehensive analysis of semi-pure proteins with minimal non-target protein contamination. Examples of good candidates for this approach include recombinantly-expressed proteins, in vitro enzyme assay experiments and the characterization of bioactive peptides.
Related Publications
Hong SH, Ntai I, Haimovich AD, Kelleher NL, Isaacs FJ, Jewett MC “Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation.” ACS Synth Biol, 2014, 3(6) p. 398-409.
Summary | Full Text (PMC) | Full Text (DOI)
Siuti N, Kelleher NL “Decoding protein modifications using top-down mass spectrometry.” Nat Methods, 2007, 4(10) p. 817-21.
Summary | Full Text (PMC) | Full Text (DOI)
Savaryn JP, Catherman AD, Thomas PM, Abecassis MM, Kelleher NL “The emergence of top-down proteomics in clinical research.” Genome Med, 2013, 5(6) p. 53.
Summary | Full Text (PMC) | Full Text (DOI)
Pricing
Northwestern/CBC: Starting at $202 (depending on number of samples submitted and length of LCMS gradient)
LCC: Starting at $184 (depending on number of samples submitted and length of LCMS gradient)
External Academic/Nonprofit: Starting at $303 (depending on number of samples submitted and length of LCMS gradient)
External For-Profit Industry: Starting at $505 (depending on number of samples submitted and length of LCMS gradient)
Instruments Used
Orbitrap Elite
Q Exactive HF
Q Exactive UHMR